Chapter 2 JAMES data formats
2.1 Objective
This chapter describes
- the format of the input child data accepted by JAMES;
- the internal format used in the code in the
Rpackages on https://github.com/growthcharts.
2.2 Input child data accepted by JAMES
The specification for the input data
- follows the definition of the Basisdataset JGZ 4.0.1;
- defines data objects;
- defines the actions taken by JAMES in case of incorrect, missing or out-of-range data;
- defines the error messages for informing the client.
Data accepted by JAMES should follow the JSON schema defined at https://raw.githubusercontent.com/growthcharts/bdsreader/srm/inst/schemas/bds_v3.0.json.
For backward compatibility, JAMES supports older versions of the schema (v1.0, v1.1 and v2.0). Do not use these deprecated formats for new applications.
2.3 Error checking policy
Error checking of the JSON data occurs in three phases:
PHASE 1: Check whether the JSON data are valid JSON. The process terminates with an error message if the input JSON is not valid.
PHASE 2: Validate the JSON data against the JSON schema specification. The process terminates with an error if any required fields are missing. The process generates messages for data points that do not conform to the JSON schema, but continues.
PHASE 3: Check the range of the numeric data. The process generates messages for out-of-range values, but continues using the specified values.
The default JSON schema in PHASE 2 is the built-in JSON schema bds_v3.0.json, a data format implementing a version that accepts strings as values for BDS-elements.
2.4 Checking structure of the input data against the JSON schema
The inst/extdata/bds_v3.0 directory of the jamesdemodata package contains examples of input data in JSON format. Non-R user can access these from GitHub from link https://github.com/growthcharts/jamesdemodata/tree/master/inst/extdata/bds_v3.0.
Manual checking the structure of your child data can be done as follows. Surf to https://www.jsonschemavalidator.net, paste the JSON schema definition in the left panel and paste the child data in the right panel. You should see something like the Figure 2.1.
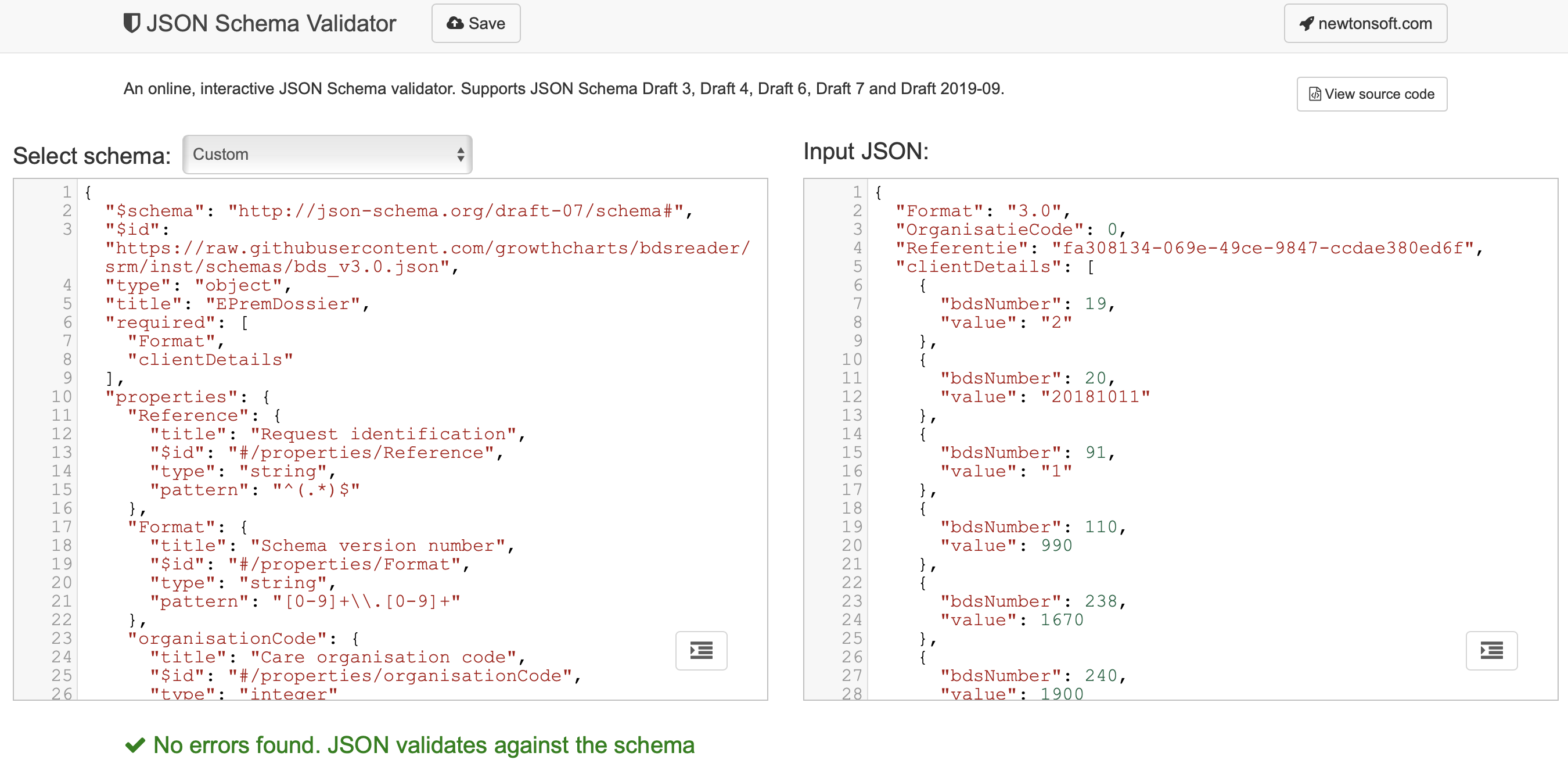
Figure 2.1: Manual validation of a child dataset (right side) according JSON schema bds_v3.0.json.
Experiment with your child file (e.g. remove the required Format field) to see what types errors the validator can catch.
2.5 BDS-elements supported by JAMES
| BDS | Description | Value | Label | R name |
|---|---|---|---|---|
| 19 | Sex of child | “0” | Unknown | sex |
| “1” | Male | |||
| “2” | Female | |||
| “3” | Not specified | |||
| 20 | Date of birth | “yyyymmdd” | year-month-day | dob |
| 62 | Caretaker relation | “01” | biological father | |
| “02” | biological mother | |||
| “03” | male partner, stepfather | |||
| “04” | female partner, stepmother | |||
| “05” | adoptive father | |||
| “06” | adoptive mother | |||
| “07” | foster father | |||
| “08” | foster mother | |||
| “98” | other | |||
| 63 | Caretaker date of birth | “yyyymmdd” | year-month-day | dobf |
dobm |
||||
agem |
||||
| 66 | Caretaker education | “01” | no primary school | - |
| “02” | primary school, special ed | |||
| “03” | VSO-MLK/IVBO/VMBO-LWOO | |||
| “04” | LBO/VBO/VMBO-BBL&KBL | |||
| “05” | MAVO/VMBO-GL&TL | |||
| “06” | MBO | |||
| “07” | HAVO/VWO | |||
| “08” | HBO/HTS/HEAO | |||
| “09” | WO | |||
| “98” | Other | |||
| “00” | Unknown | |||
| 71 | Caretaker birth country | “dddd” | 4-digit code, Table 34 | etn (always NL) |
| 82 | Gestational age | “ddd” | in days | gad |
| 91 | Smoking during pregnancy | “1” | yes | smo |
| “2” | no | |||
| “99” | unknown | |||
| 110 | Birth weight | “dddd” | 3-4 digits, grammes | bw (g) |
| 235 | Length/height | “dddd” | 3-4 digits, millimeters | hgt (cm) |
| 245 | Body weight | “dddddd” | 3-6 digits, grammes | wgt (kg) |
| 252 | Head circumference | “ddd” | 2-3 digits, millimeters | hdc (cm) |
| 238 | Height biological mother | “dddd” | 3-4 digits, millimeters | hgtf (cm) |
| 240 | Height biological father | “dddd” | 3-4 digits, millimeters | hgtm (cm) |
| 510 | Passive smoking | “01” | No smoking in house | - |
| “02” | Never with child | |||
| “03” | Not in last 7 days | |||
| “04” | Yes |
JAMES supports the following BDS numbers with Van Wiechen items: 879, 881, 883, 884, 885, 887, 888, 889, 890, 891, 894, 896, 897, 898, 902, 903, 906, 907, 910, 912, 914, 916, 917, 918, 920, 922, 923, 926, 945, 951, 955, 956, 958, 959, 961, 962, 964, 966, 968, 970, 971, 973, 975, 977, 978, 986, 989, 991, 993, 994, 996, 999, 1002, 886, 892, 893, 900, 905, 909, 913, 921, 927, 928, 930, 931, 932, 933, 934, 935, 936, 937, 938, 939, 940, 941, 943, 947, 948, 949, 950, 953, 954, 972, 980, 982, 984, 998, 1001, 1278.
The results of the Van Wiechen items are converted into 0/1 codes (by bdsreader:::convert_ddi_gsed) and stored with GSED 9-position names as defined by the dscore package.
In addition, JAMES reads the following non-BDS fields:
| Fields | Description | Type | R name |
|---|---|---|---|
| Reference | Description (opt) | String | name |
| Format | JSON schema number (req) | Number.Number | - |
| organisationCode | Organisation code (opt) | Integer | src |
2.6 JAMES internal data
Suppose we coded the following data set.
{
"Format": "3.0",
"organisationCode": 1234,
"Reference": "Maria",
"clientDetails": [
{
"bdsNumber": 19,
"value": "2"
},
{
"bdsNumber": 20,
"value": "20181011"
},
{
"bdsNumber": 82,
"value": 189
},
{
"bdsNumber": 91,
"value": "1"
},
{
"bdsNumber": 110,
"value": 990
},
{
"bdsNumber": 238,
"value": 1670
},
{
"bdsNumber": 240,
"value": 1900
}
],
"clientMeasurements": [
{
"bdsNumber": 235,
"values": [
{
"date": "20181111",
"value": 380
},
{
"date": "20181211",
"value": 435
}
]
},
{
"bdsNumber": 245,
"values": [
{
"date": "20181011",
"value": 990
},
{
"date": "20181111",
"value": 1250
},
{
"date": "20181211",
"value": 2100
}
]
},
{
"bdsNumber": 252,
"values": [
{
"date": "20181111",
"value": 270
},
{
"date": "20181211",
"value": 305
}
]
}
],
"nestedDetails": [
{
"nestingBdsNumber": 62,
"nestingCode": "01",
"clientDetails": [
{
"bdsNumber": 63,
"value": "19950704"
}
],
"clientMeasurements": [
]
},
{
"nestingBdsNumber": 62,
"nestingCode": "02",
"clientDetails": [
{
"bdsNumber": 63,
"value": "19901202"
}
],
"clientMeasurements": [
]
}
]
}The following R script shows reading and conversion of the data.
library(bdsreader)
fn <- system.file("examples/maria.json", package = "bdsreader")
m <- read_bds(fn)Object m object is a list with two components:
m$psna tibble with one row containing fixed covariatesm$xyza tibble with multiple rows with time-varying data
m$psn## # A tibble: 1 × 16
## id name dob dobf dobm src dnr sex gad ga
## <int> <chr> <date> <date> <date> <chr> <chr> <chr> <dbl> <dbl>
## 1 -1 Maria 2018-10-11 1995-07-04 1990-12-02 1234 <NA> female 189 27
## # ℹ 6 more variables: smo <int>, bw <dbl>, hgtm <dbl>, hgtf <dbl>, agem <dbl>,
## # etn <chr>m$xyz## # A tibble: 11 × 8
## age xname yname zname zref x y z
## <dbl> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 0.0849 age hgt hgt_z nl_2012_hgt_female_27 0.0849 38 -0.158
## 2 0.0849 age wgt wgt_z nl_2012_wgt_female_27 0.0849 1.25 -0.203
## 3 0.0849 age hdc hdc_z nl_2012_hdc_female_27 0.0849 27 -0.709
## 4 0.0849 age bmi bmi_z nl_1997_bmi_female_nl 0.0849 8.66 -5.72
## 5 0.167 age hgt hgt_z nl_2012_hgt_female_27 0.167 43.5 0.047
## 6 0.167 age wgt wgt_z nl_2012_wgt_female_27 0.167 2.1 0.015
## 7 0.167 age hdc hdc_z nl_2012_hdc_female_27 0.167 30.5 -0.913
## 8 0.167 age bmi bmi_z nl_1997_bmi_female_nl 0.167 11.1 -3.77
## 9 0 age wgt wgt_z nl_2012_wgt_female_27 0 0.99 0.19
## 10 0.0849 hgt wfh wfh_z nl_2012_wfh_female_ 38 1.25 -0.001
## 11 0.167 hgt wfh wfh_z nl_2012_wfh_female_ 43.5 2.1 0.326